Tutorial
Exploring diel carbon flux cycles
Authors: [David Durden]
Last Updated: Apr 20, 2023
This data tutorial provides an overview of exploring NEON carbon
flux data, using the neonUtilities R package. If you are just starting to work with NEON data it is recommended that you also checkout the more general tutorials, such as the
neonUtilities tutorial
or the Introduction to working with NEON eddy flux data for a more flux centric introductory tutorial. This tutorial builds off the data exploration techniques described in the aforementioned tutorials, and will explore carbon dynamics at two forested ecosystem sites with very different land management practices in NEON's Domain 5 - Great Lakes, STEI and TREE. The STEI and TREE sites are only located approximately 1 mile from each, so the observed differences in fluxes can be attributed to land management practices associated with the forests.
This tutorial assumes general familiarity with eddy-covariance data, and will briefly touch discuss both turbulent (turb) and storage (stor) fluxes in relation to the net surface atmosphere exchange (nsae). The nsae for CO2 fluxes often referred to as Net Ecosystem Exchange (NEE).
1. Setup
Start by installing and loading packages and setting options. The function below will evaluate if the required packages (packReq) already exist in you R library, and will install and load these packages. The rhdf5 package is hosted on Bioconductor requiring a BiocManager to be installed to allow rhdf5 to be pulled from the Bioconductor repository:
#Required R packages
packReq <- c("BiocManager", "rhdf5", 'neonUtilities', 'ggplot2','tidyverse', "lubridate")
#Install and load all required packages
lapply(packReq, function(x) {
if(require(x, character.only = TRUE) == FALSE) {
install.packages(x)
library(x, character.only = TRUE)
}})
options(stringsAsFactors=F)
Now that all the required packages are loaded, let's define all the necessary function arguments to download and stack our flux data using the neonUtilities package's zipsByProduct() and stackEddy() functions. I will define the input arguments as variables to make it easy to modify if I want to explore additional sites or months of data. For this analysis, we will focus on the 2022 growing season at STEI and TREE. Please be aware that the data download size is ~ 1 GB. The code can be modified to shorten or extend this window, but to get a good look at the carbon dynamics at the site it was important to capture most of the growing season.
Inputs to the zipsByProduct() function:
-
dpID: DP4.00200.001, the bundled eddy covariance product -
package: basic (the expanded package has additional quality metrics and advanced footprint matrices that are not relevant to this tutoiral) -
site: STEI and TREE -
startate: 2022-04 -
enddate: 2022-09 -
savepath: modify this to something logical on your machine or use the Rtempdir() -
check.size: TRUE if you want to see file size before downloading, otherwise FALSE
The download may take a while, especially if you're on a slow network. For faster downloads, consider using an API token.
#Target dates
startDate <- "2022-04"
endDate <- "2022-09"
#Site
site <- c("STEI", "TREE")
#File directory
dirFile <- c(tempdir(),"/home/ddurden/eddy/tmp/tutorial")[2]
zipsByProduct(dpID="DP4.00200.001", package="basic",
site=site,
startdate=startDate, enddate=endDate,
savepath=dirFile,
check.size=FALSE)
2. Stacking Level 4 Flux Data
There are five levels of data contained in the eddy flux bundle. For full details, refer to the .
In this tutorial we will only be focusing on Level 4 (dp04) flux data products; however, additional data products from Level 0' (dp0p) calibrated raw data to Level 3 (dp03) spatially interpolated vertical profiles used to derive the storage flux are available in the EC bundled HDF5 files. Information can be found on the webpage. The Introduction to working with NEON eddy flux data tutorial dives into additional detail regarding the other data product levels, but this tutoiral will focus exclusively on flux data.
To extract the dp04 data from the HDF5 files and merge them into a
single table, we'll use the stackEddy() function. We provide the function to input arguments, filepath and level. The filepath will be the file directory (dirFile) used for the data download via zipsByProduct() combined with the filestoStack00200 folder created by the function. To grab just the flux data products level = dp04:
flux <- neonUtilities::stackEddy(filepath=paste0(dirFile,"/filesToStack00200"),
level="dp04")
We now have an object called flux. It's a named list containing four
tables: one table for each site's data, and variables and objDesc
tables. One of the advantages to the HDF5 files is all the metadata and data descriptions can be compiled into file as additional data tables or as attributes. The stackEddy() function stacks this contextual data in the variables and objDesc tables to help you interpret the column headers in the data table (though it's not comprehensive currently).
The terms of interest for our analysis in dp04 the three flux quantities under
fluxCo2:
-
turb: Turbulent flux -
stor: Storage -
nsae: Net surface-atmosphere exchange
Note the variables table contains the units for each field:
flux$variables
## category system variable stat units
## 1 data fluxCo2 nsae timeBgn NA
## 2 data fluxCo2 nsae timeEnd NA
## 3 data fluxCo2 nsae flux umolCo2 m-2 s-1
## 4 data fluxCo2 stor timeBgn NA
## 5 data fluxCo2 stor timeEnd NA
## 6 data fluxCo2 stor flux umolCo2 m-2 s-1
## 7 data fluxCo2 turb timeBgn NA
## 8 data fluxCo2 turb timeEnd NA
## 9 data fluxCo2 turb flux umolCo2 m-2 s-1
## 10 data fluxH2o nsae timeBgn NA
## 11 data fluxH2o nsae timeEnd NA
## 12 data fluxH2o nsae flux W m-2
## 13 data fluxH2o stor timeBgn NA
## 14 data fluxH2o stor timeEnd NA
## 15 data fluxH2o stor flux W m-2
## 16 data fluxH2o turb timeBgn NA
## 17 data fluxH2o turb timeEnd NA
## 18 data fluxH2o turb flux W m-2
## 19 data fluxMome turb timeBgn NA
## 20 data fluxMome turb timeEnd NA
## 21 data fluxMome turb veloFric m s-1
## 22 data fluxTemp nsae timeBgn NA
## 23 data fluxTemp nsae timeEnd NA
## 24 data fluxTemp nsae flux W m-2
## 25 data fluxTemp stor timeBgn NA
## 26 data fluxTemp stor timeEnd NA
## 27 data fluxTemp stor flux W m-2
## 28 data fluxTemp turb timeBgn NA
## 29 data fluxTemp turb timeEnd NA
## 30 data fluxTemp turb flux W m-2
## 31 data foot stat timeBgn NA
## 32 data foot stat timeEnd NA
## 33 data foot stat angZaxsErth deg
## 34 data foot stat distReso m
## 35 data foot stat veloYaxsHorSd m s-1
## 36 data foot stat veloZaxsHorSd m s-1
## 37 data foot stat veloFric m s-1
## 38 data foot stat distZaxsMeasDisp m
## 39 data foot stat distZaxsRgh m
## 40 data foot stat distZaxsAbl m
## 41 data foot stat distXaxs90 m
## 42 data foot stat distXaxsMax m
## 43 data foot stat distYaxs90 m
## 44 qfqm fluxCo2 nsae timeBgn NA
## 45 qfqm fluxCo2 nsae timeEnd NA
## 46 qfqm fluxCo2 nsae qfFinl NA
## 47 qfqm fluxCo2 stor qfFinl NA
## 48 qfqm fluxCo2 stor timeBgn NA
## 49 qfqm fluxCo2 stor timeEnd NA
## 50 qfqm fluxCo2 turb timeBgn NA
## 51 qfqm fluxCo2 turb timeEnd NA
## 52 qfqm fluxCo2 turb qfFinl NA
## 53 qfqm fluxH2o nsae timeBgn NA
## 54 qfqm fluxH2o nsae timeEnd NA
## 55 qfqm fluxH2o nsae qfFinl NA
## 56 qfqm fluxH2o stor qfFinl NA
## 57 qfqm fluxH2o stor timeBgn NA
## 58 qfqm fluxH2o stor timeEnd NA
## 59 qfqm fluxH2o turb timeBgn NA
## 60 qfqm fluxH2o turb timeEnd NA
## 61 qfqm fluxH2o turb qfFinl NA
## 62 qfqm fluxMome turb timeBgn NA
## 63 qfqm fluxMome turb timeEnd NA
## 64 qfqm fluxMome turb qfFinl NA
## 65 qfqm fluxTemp nsae timeBgn NA
## 66 qfqm fluxTemp nsae timeEnd NA
## 67 qfqm fluxTemp nsae qfFinl NA
## 68 qfqm fluxTemp stor qfFinl NA
## 69 qfqm fluxTemp stor timeBgn NA
## 70 qfqm fluxTemp stor timeEnd NA
## 71 qfqm fluxTemp turb timeBgn NA
## 72 qfqm fluxTemp turb timeEnd NA
## 73 qfqm fluxTemp turb qfFinl NA
## 74 qfqm foot turb timeBgn NA
## 75 qfqm foot turb timeEnd NA
## 76 qfqm foot turb qfFinl NA
3. Plotting CO2 Flux Data
Let's look at some data! First, we will combine the data from the two sites into a single dataframe with an additional factor variable for the site to allow us to utilize tidyverse and ggplot2 packages for some data munging and to create some interesting data exploration visualizations. In this tutorial we utilize the piping operater %>% from the dplyr to allow us to quickly subset, filter, sort, and plot data using just our combined dataframe dfFlux. We start by looking at the turbulent CO2 flux timeseries at TREE with the points colored by qfFinl:
flux$STEI$Site <- "STEI"
flux$TREE$Site <- "TREE"
dfFlux <- rbind.data.frame(flux$STEI, flux$TREE)
dfFlux$Site <- as.factor(dfFlux$Site)
dfFlux %>%
ggplot(aes(timeBgn, data.fluxCo2.turb.flux, color=factor(qfqm.fluxCo2.turb.qfFinl) )) +
geom_point() +
scale_color_brewer(palette="Set2", name="qfFinal") +
facet_grid(~Site)
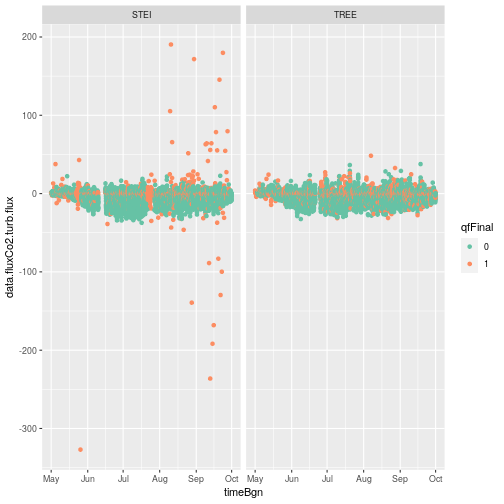
When we plot the data turbulent CO2 flux data, we see a good agreement
between the STEI and TREE sites. We also see some outliers that look to be due to
erroneous data, especially at the STEI site. Fortunately, it looks like final
quality flag (qfFinal = qfFinl) caught the largest outliers, and we can use it to remove
these data from the analysis. Let's first dig into a little background information
about NEON bundled EC quality flags.
Quality flags
The NEON Surface-atmosphere exchange (SAE) data products all come accompanied by
a qfFinal. Information the defining quality flag (qf), quality metric (qm),
and the framework to derive qfFinal are detailed in and
. Ultimately, for the dp04 flux data
products (e.g. data.fluxCo2.turb.flux), qfFinal of dp01 constituent data
streams derived from plausibility tests (i.e. range test, step test, spike test,
and persistence test) and sensor diagnostic flags, fluxCo2 = rtioMoleDryCo2
and veloZaxsErth (dp01 data outside the scope of tutorial), are propagated and
combined with theoretical eddy-covariance specific quality tests (i.e., stationarity,
integral turbulence characteristics tests) to determine whether the data product
is flagged as valid (qfFinal = 0) or invalid (qfFinal = 1).
Let's summarize our qfqm.fluxCo2.turb.qfFinl to see the total flagged percentage
at our 2 sites.
dfFlux %>% group_by(Site) %>% summarise(mean(qfqm.fluxCo2.turb.qfFinl))
## # A tibble: 2 x 2
## Site `mean(qfqm.fluxCo2.turb.qfFinl)`
## <fct> <dbl>
## 1 STEI 0.219
## 2 TREE 0.261
Now, lets plot the qfFinal failed percentage for all our flux data products:
dfFlux %>%
select(contains("qfqm")) %>%
pivot_longer(cols = everything(), names_to = "var") %>%
group_by(var) %>%
summarise(mean_var = mean(value) * 100) %>%
ggplot(aes(x = var, y = mean_var, fill = var)) +
geom_col() +
guides(x = guide_axis(angle = 90)) +
labs(x="Variable", y="Percent qfFinal failed") +
scale_fill_brewer(palette="RdYlBu")
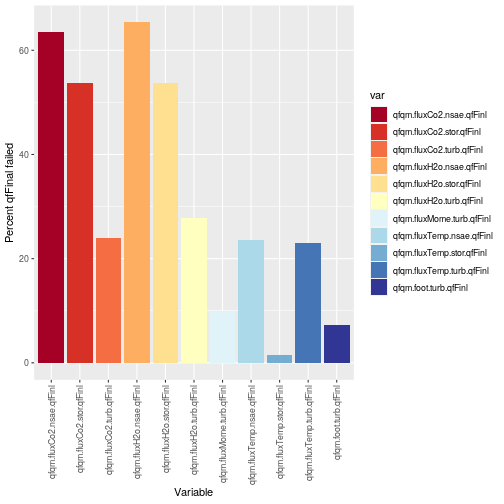
Removing data with failed qfFinal
We see that qfqm.fluxCo2.turb.qfFinl is flagged ~ 20-25% over the growing season; however,
the qfqm.fluxCo2.stor.qfFinl and qfqm.fluxCo2.nsae.qfFinl are flagged quite a
bit more due to the complexity of the system and propagation of issues between
sub-components to the overall system (additionally, we currently employ a conservative
quality control for the storage fluxes). Now that we have evaluated the qfFinal
for all the fluxCo2 data streams, lets remove flagged data from our dataframe.
dfFlux %>% select(contains("qfqm") & contains("fluxCO2")) %>% summarise_each(sum)
## qfqm.fluxCo2.nsae.qfFinl qfqm.fluxCo2.stor.qfFinl qfqm.fluxCo2.turb.qfFinl
## 1 9322 7886 3526
dfFlux %>% select(contains("data") & contains("fluxCO2")) %>% summarise_each(funs(sum(is.na(.))))
## data.fluxCo2.nsae.flux data.fluxCo2.stor.flux data.fluxCo2.turb.flux
## 1 3174 2334 1366
dfFlux$data.fluxCo2.turb.flux[(which(dfFlux$qfqm.fluxCo2.turb.qfFinl== 1))] <- NaN
dfFlux$data.fluxCo2.stor.flux[(which(dfFlux$qfqm.fluxCo2.stor.qfFinl== 1))] <- NaN
dfFlux$data.fluxCo2.nsae.flux[(which(dfFlux$qfqm.fluxCo2.nsae.qfFinl== 1))] <- NaN
dfFlux %>% select(contains("qfqm") & contains("fluxCO2")) %>% summarise_each(sum)
## qfqm.fluxCo2.nsae.qfFinl qfqm.fluxCo2.stor.qfFinl qfqm.fluxCo2.turb.qfFinl
## 1 9322 7886 3526
dfFlux %>% select(contains("data") & contains("fluxCO2")) %>% summarise_each(funs(sum(is.na(.))))
## data.fluxCo2.nsae.flux data.fluxCo2.stor.flux data.fluxCo2.turb.flux
## 1 9713 7913 4343
We see from the summary of the fluxCo2 qfFinal and data NA's, that we have
removed all the data from data.fluxCo2.turb.flux, data.fluxCo2.turb.flux,
and data.fluxCo2.turb.flux. Now we have a clean data set to begin our diel
flux cycle data analysis.
3. Plotting the diel cycle of CO2 fluxes
Let's look at the data in a different way, the diel cycle of carbon and water
vapor are important when evaluating carbon sequestration and ecosystem health.
We can evaluate the diel cycle by binning the data by hour in the day, this is
relatively straightforward using ggplot and the boxplot function (geom_boxplot()).
dfFlux$hour <- factor(lubridate::hour(dfFlux$timeBgn))
ggplot(dfFlux, aes(x = hour, y = data.fluxCo2.turb.flux, fill = Site)) +
geom_boxplot() +
stat_summary(fun = median, geom = 'line', aes(group = Site, colour = Site))
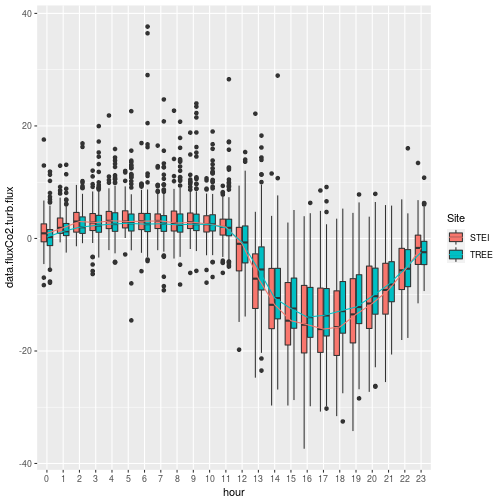
Nice! We get a glimpse of daily carbon dynamics, but something doesn't quite look right.
Note the timing of C uptake; the UTC time zone is clear here, where
uptake occurs at times that appear to be during the night. Note that
all NEON data use UTC time, aka Greenwich Mean Time.
This is true across NEON's instrumented, observational, and airborne
measurements. When working with NEON data, it's best to keep
everything in UTC as much as possible, otherwise it's very easy to
end up with data in mismatched times. We may want to convert the time to Local
standard time (LST). The metadata attributes at the site level in the bundled
EC HDF5 files contains fields for ZoneTime and TimeDiffUtcLt. We can use the
rhdf5 package h5readAttributes() function to grab the metadata. In this
analysis, both the STEI and TREE are in the same timezone. Therefore, we will
only grab metadata from one file and apply to both sites.
fileMeta <- list.files(dirFile, pattern = paste0(".*",site[1],".*.h5"), recursive = TRUE, full.names = TRUE)[1]
siteMeta <- h5readAttributes(fileMeta, name = site[1])
siteMeta
## $DistZaxsCnpy
## [1] 9.5
##
## $DistZaxsDisp
## [1] "6.2"
##
## $DistZaxsGrndOfst
## [1] "0.758"
##
## $DistZaxsLvlMeasTow
## [1] "0.65" "4.79" "8.15" "12.30" "15.66" "22.36"
##
## $DistZaxsTow
## [1] "21.34"
##
## $ElevRefeTow
## [1] "475.7317"
##
## $LatTow
## [1] 45.50895
##
## $LonTow
## [1] -89.58636
##
## $LvlMeasTow
## [1] 6
##
## $`Pf$AngEnuXaxs`
## [1] 0.01544
##
## $`Pf$AngEnuYaxs`
## [1] -0.019366
##
## $`Pf$Ofst`
## [1] 0.05764
##
## $TimeDiffUtcLt
## [1] -6
##
## $TimeTube
## [1] 10.1 11.1 10.4 11.3 11.3 12.2
##
## $TypeEco
## [1] "Aspen dominated regenerating forest"
##
## $ZoneTime
## [1] "CST"
##
## $ZoneUtm
## [1] "16"
dfFlux$timeBgnLst <- dfFlux$timeBgn + lubridate::hours(siteMeta$TimeDiffUtcLt)
dfFlux$hourLst <- factor(lubridate::hour(dfFlux$timeBgnLst))
ggplot(dfFlux, aes(x = hourLst, y = data.fluxCo2.turb.flux, fill = Site)) +
geom_boxplot() +
stat_summary(fun = median, geom = 'line', aes(group = Site, colour = Site)) +
scale_y_continuous(limits = quantile(dfFlux$data.fluxCo2.turb.flux, c(0.001, 0.999), na.rm = TRUE))
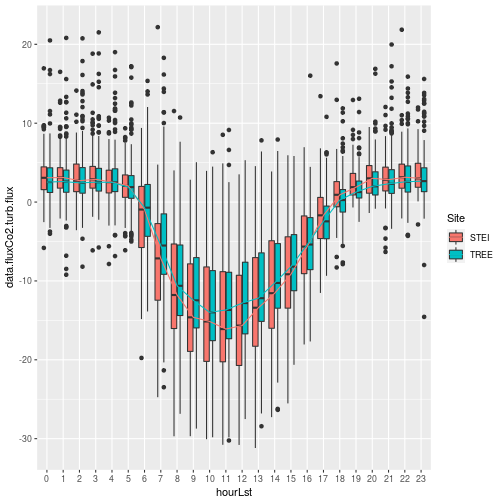
That looks better, now we can clearly see the diel cycle of the turbulent
CO2 flux at our STEI and TREE sites with peak carbon uptake around noon.
The carbon uptake should align with peak solar angle and incoming
photosynthetically active radiation (PAR). The boxplot also provides us information
about the variability of the fluxes throughout the day across the growing season
with interquartile range (IQR) represented by the box and the whiskers indicating
the threshold for outliers (black dots) as 1.5 * IQR subtracted from 1st quantile (Q1)
and added to 3rd quantile (Q3):
Q1 - 1.5 * IQR
Q3 + 1.5 * IQR
In the boxplots above the outlier points were greatly reduced after the qfFinal
data removal; however, some outliers remained. To focus our attention on the
bulk of the data, we scaled the y-axis using scale_y_continuous() in conjunction
with the quantile() function to look at 99.8% of the data excluding extreme
outliers that may have remained after our filtering.
We know that the turbulent flux is usually the dominant flux component to NEE,
but the storage flux can be significant at forested sites. Let's have a look at
the storage flux and it's impact on the NSAE flux at STEI and TREE:
ggplot(dfFlux, aes(x = hourLst, y = data.fluxCo2.stor.flux, fill = Site)) +
geom_boxplot() +
stat_summary(fun = median, geom = 'line', aes(group = Site, colour = Site)) +
scale_y_continuous(limits = quantile(dfFlux$data.fluxCo2.stor.flux, c(0.001, 0.999), na.rm = TRUE))
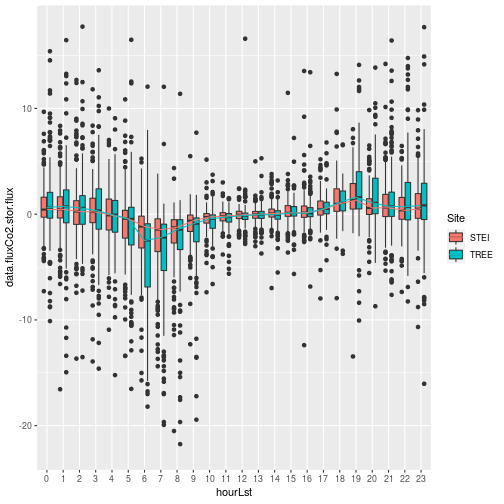
The storage flux as expected is quite smaller than the turbulent flux, and is dominant during the nighttime hours and evening/morning transition times when turbulence is generally weaker and the trees and understory vegetation aren't photosynthesizing. It's interesting to note the stronger stronger signal at the TREE site.Let's look how that impacts the overall NSAE flux:
ggplot(dfFlux, aes(x = hourLst, y = data.fluxCo2.nsae.flux, fill = Site)) +
geom_boxplot() +
stat_summary(fun = median, geom = 'line', aes(group = Site, colour = Site)) +
scale_y_continuous(limits = quantile(dfFlux$data.fluxCo2.nsae.flux, c(0.001, 0.999), na.rm = TRUE))
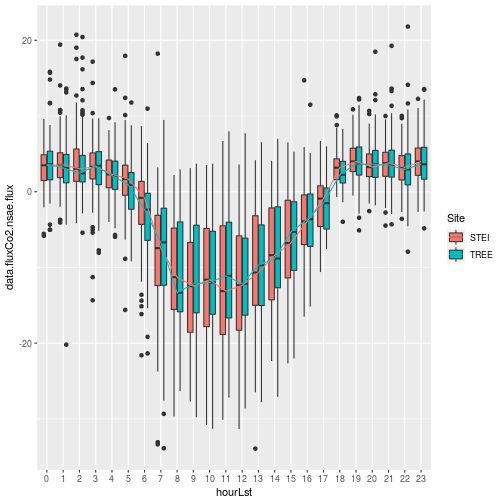
When we look at the NSAE flux we see that the STEI site appears to sequester more carbon, which makes sense considering the STEI site is predominately a young Aspen stand for pulpwood that is growing at a fast rate. It's difficult to get a good total Carbon uptake estimate without performing some more advanced gap-filling and partitioning to better understand the ecosystem carbon dynamics. We can simply look at the relative cumulative flux contributions:
dfFlux %>%
select(contains("data.fluxCo2") | "Site") %>%
group_by(Site) %>%
summarise_each(funs(mean(., na.rm = TRUE)))
## # A tibble: 2 x 4
## Site data.fluxCo2.nsae.flux data.fluxCo2.stor.flux data.fluxCo2.turb.flux
## <fct> <dbl> <dbl> <dbl>
## 1 STEI -2.72 -0.0259 -3.83
## 2 TREE -2.54 -0.0911 -3.14
As expected from the plots, we see that on average the STEI site takes up more
carbon on average across the growing season when we look at nsae and turb
CO2 fluxes. This obviously comes with the tradeoff that the trees are
managed for cultivation in the future; whereas, TREE is currently managed as
an educational resource that is likely to be preserved, which has other ecosystem
health benefits.
This analysis is meant to provide a glimpse into NEON flux data products, while focusing primarily on CO2. This workflow can easily be adapted to look at Sensible and Latent heat (H2O) fluxes, or be adapted to evaluate additional NEON sites or years of data.